Metagenomic Next-Generation Sequencing for Infectious Disease Diagnosis
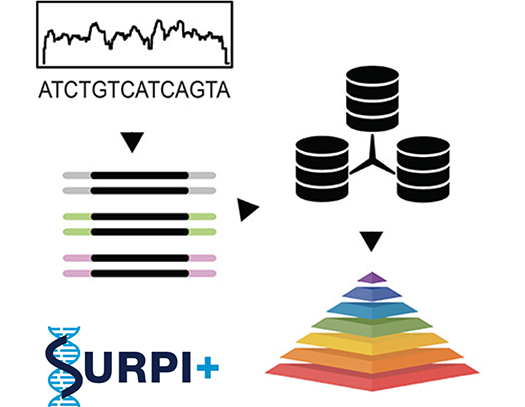
We are clinically validating metagenomic next-generation sequencing (mNGS) assays for infectious disease diagnosis in a CLIA-certified laboratory. This effort has been spearheaded by the Precision Diagnosis of Acute Infectious Diseases study, sponsored by the California Initiative to Advance Precision Medicine. Clinical testing for patients is now available. This work was highlighted in TIME magazine in May 2017 as a promising approach to investigating pandemics.
Zika Virus
The Chiu lab is working to further the understanding of Zika virus by studying its ability to infect brain and spinal cord cells as well as identifying and interpreting mutations that arise during the course of infection, especially during pregnancies. We are also actively collaborating with other groups (Texas Biomedical Research Institute, UC Davis, etc.) to develop nonhuman primate models of Zika virus infection.
Host Response to Infection
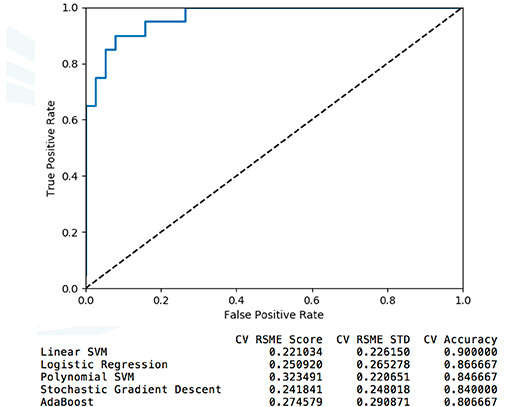
We are interested in leveraging RNA-seq (transcriptome) approaches to understand the human host response to infection and use this information for diagnostic purposes. We are generating machine (deep) learning models from RNA-seq data to either use directly for prediction or to identify biomarkers of disease.
Human Pegivirus 2
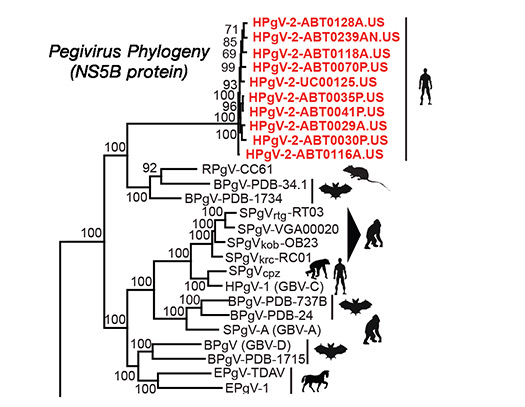
The Chiu lab co-discovered the novel virus HPgV-2 by metagenomic next-generation sequencing (mNGS) in 2015. We are now developing methods to cultivate the virus in vitro for investigations of viral infectivity and pathogenesis.
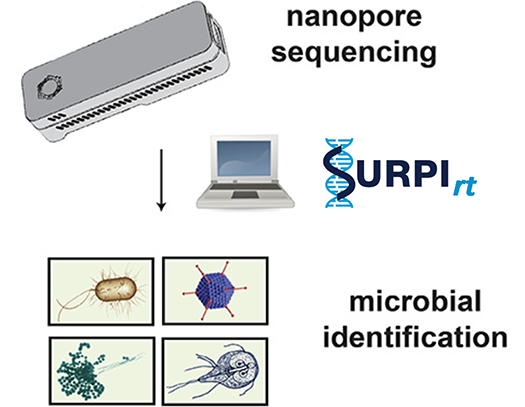
Disease Surveillance and Diagnosis by Nanopore Sequencing
The nanopore sequencing platform on the MinION and GridION instruments from Oxford Nanopore Technologies offers an opportunity to sequence DNA in real time. We are working to optimize protocols for this tool so that it can be deployed during outbreaks in remote field settings or validated as a diagnostic sequencing assay in the clinical laboratory.
Transfusion-Transmitted Diseases
Researchers in the Chiu lab are identifying bloodborne pathogens that can cause disease in blood transfusion recipients. We are also developing techniques to improve methods for screening blood donations for known infectious agents.
Virome / Microbiome in Human Health
As the impact of the microbiome on human health is becoming more apparent, we are interested in characterizing the virome (as well as bacterial microbiome) and how it develops in early life. The Chiu lab collaborates with Dr. Julie Parsonnet at Stanford University on the STORK project, an ‘omics study of a longitudinal birth cohort in Northern California from ages 0 to 2 years.
Sequencing in Space
Through our partnership with NASA and the Mason laboratory at Weill-Cornell Medical Center, we helped make sequencing in space a reality. We are now working to expand the sequencing capabilities in the international space station and for future cis-lunar and Mars missions, and to create tools to allow for real-time data analysis onboard.
Image Credit: NASA
Tickborne Diseases
Lyme and other tickborne diseases are a major public health concern in the U.S., but remain difficult to diagnose. We are interested in improving the ability to accurately diagnose these diseases, both by screening directly for the pathogens themselves as well as identifying host gene expression signatures indicative of infection.
Image Credit: CDC
HIV
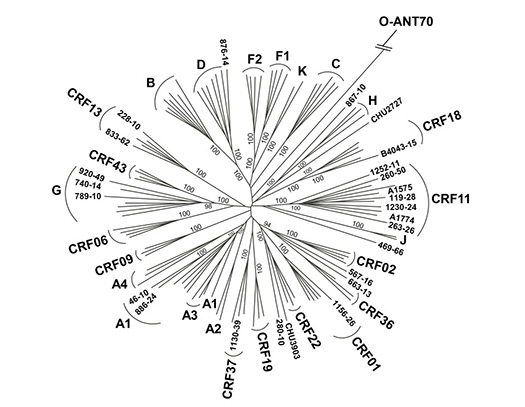
In collaboration with Abbott Laboratories, we are performing whole-genome sequencing of a number of transfusion-transmitted viruses (HIV, HBV, and HCV) to understand evolution, variation, and spread of these pathogens on a global scale.
Bioinformatic Methods Development
We develop computational pipelines such as SURPI+ / SURPIrt for rapid identification of pathogens from raw sequencing data and novel methods such as SPEMS (spiked primer enriched metagenomic sequencing) to enrich for target pathogens.
Enterovirus D-68
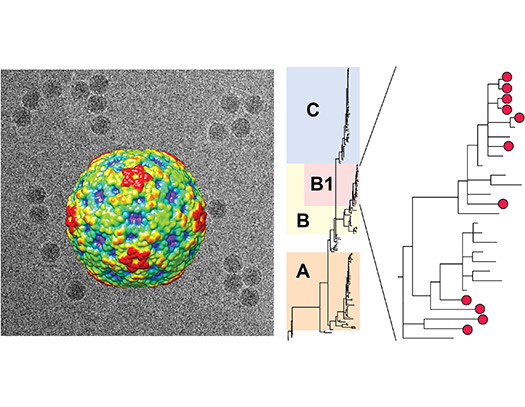
Enterovirus D-68 is a respiratory virus associated with a 2014 outbreak of acute flaccid myelitis (AFM) – paralysis of the arms, legs, and/or body from spinal cord inflammation. We showed that the majority of AFM cases in 2012-2014 were from a novel strain, named clade B1 and are actively collaborating on exploring a mouse model of EV-D68 infection.